Research
Nucleosome interactions control chromatin biology
The packaging of DNA into chromatin represents one of the most fundamental layers of cell biology. Chromatin provides the required structural compaction of the DNA to fit in the nucleus, and plays crucial roles in controlling cell fate and genomic integrity. These functions ultimately depend on the interactions of a wide range of proteins with the nucleosome, chromatin’s fundamental building block. But how these proteins recognize, bind and perturb nucleosomes to alter the functional chromatin state? Answers to these questions are vital to understand essential processes like gene regulation and DNA repair. Also, it will aid the development of novel approaches to control the epigenetic state of the cell, for example for cancer treatment or regenerative therapies. Our lab aims to resolve the underlying mechanisms of gene regulation and DNA repair, focussing at the level of nucleosome-protein interactions.
Developing Chromatin NMR Approaches
The nucleosome is a 200,000 Da supramolecular assembly of roughly one part DNA and one part protein, the histones. The massive size of the nucleosome calls for state-of-the-art NMR techniques that are tailored for such high-molecular weight systems. These can be applied both in solution and in a sediment. In addition, the histone proteins are rich in intrinsically disordered regions, the histone tails that are key in marking the functiona state of chromatin thorugh post-translational modifications.
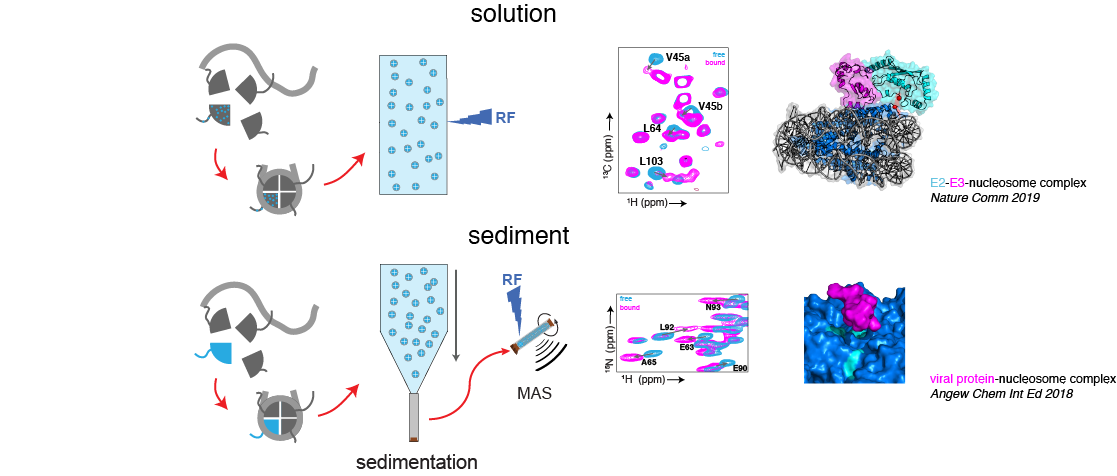
In solution, the methyl-TROSY approach allows the ultra-sensitive observation of methyl-groups in the proteins to produce beautiful high-quality spectra of the histone core. In addition, regular backbone amide based NMR can be used to study the flexible histone tails. In the sediment, we showed together with our collegues of the solid-state NMR team that 1H-detected solid-state NMR can give high quality spectra of the nucleosome core. In both cases, the NMR signals act as molecular probes to monitor the structure, dynamics and interactions of the nucleosome at atomic resolution.
We are mostly interested in the interactions of histone proteins and nucleosomes with other chromatin factors such as chaperones, remodelers or proteins that control epigenetics. The challenging nature of these systems makes them perfect for NMR-driven integrative structural biology. With the unique sensitivity of NMR spectroscopy to structure and dynamics, we hope to create a new perspective on protein-nucleosome complexes.
NMR
As the saying goes:
In theory there is no difference between theory and practice.
In practice, this is only true for NMR.
Well, at least practically true.
NMR is essentially an applied form of quantum mechanics, allowing one to accurately design and simulate experiments. The beauty of NMR is that it gives, as one of PhD’s put it, an insider’s view into the molecule of interest. The chemical shift gives NMR its extreme chemical resolving power, as if one would put a piece of cheese on the moon and you’d be able to tell whether it is jonge, belegen or oude cheese. The weak coupling of the nuclear spin to its surrounding gives the NMR signal a long life time, enabling the experimenter to control the fate of the signal almost at will and to encode structural or dynamical information into the signal. The ultimate decay of the NMR signal depends on the molecular dynamics, thus providing an atomic window into the internal motions of proteins.
